ENERGY MINIMIZATION OF NUCLEIC ACID STRUCTURE
This tutorial covers running energy minimization of nucleic-acid structures with the NARES-2P force field.
To start click MIN in the nucleic acid section (on the right, as shown in the picture below).
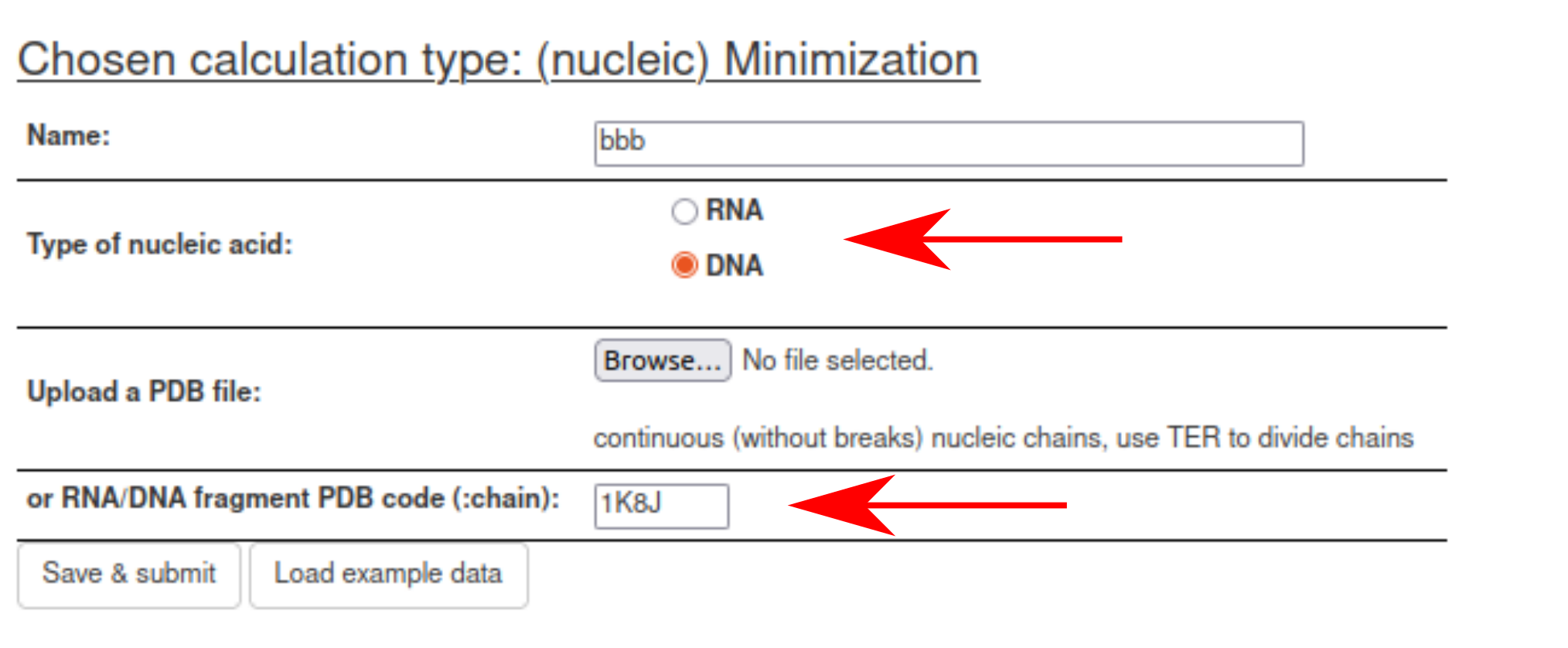
Next, upload the PDB file with the initial structure or specify the PDB code to download the structure from the PDB. Select nucleic-acid type (DNA or RNA).
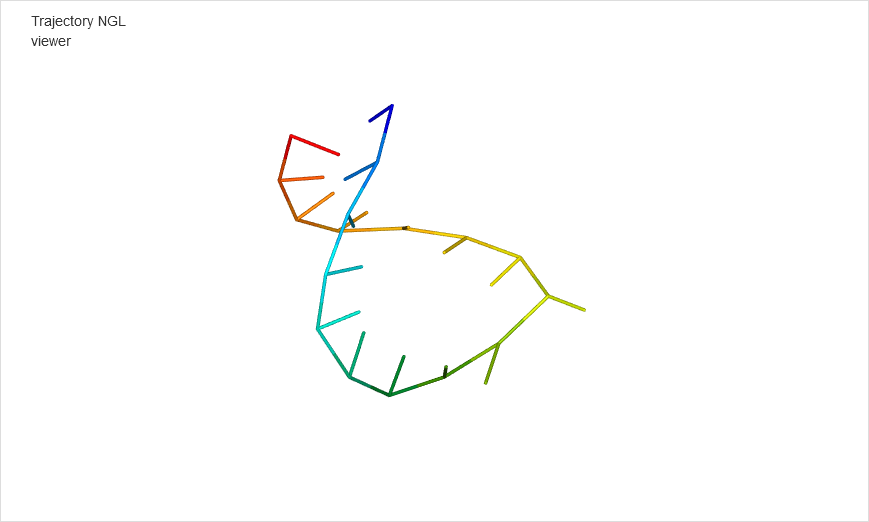
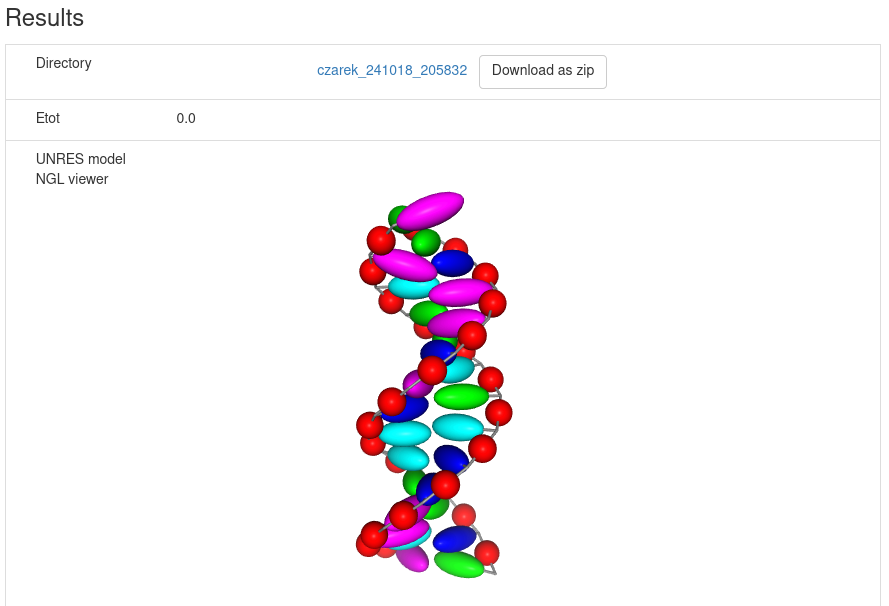
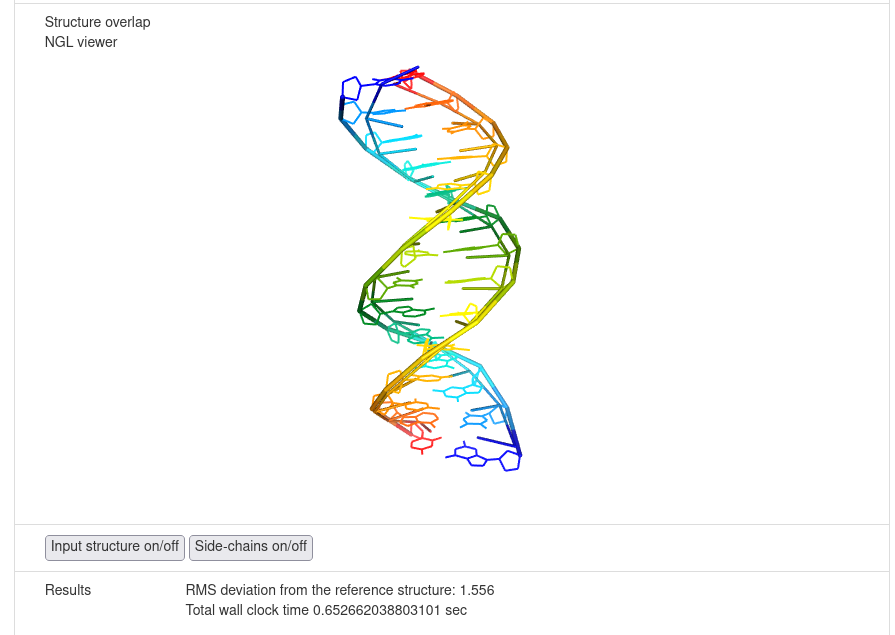
After minimization is finished, the results are displayed in the graphical form.
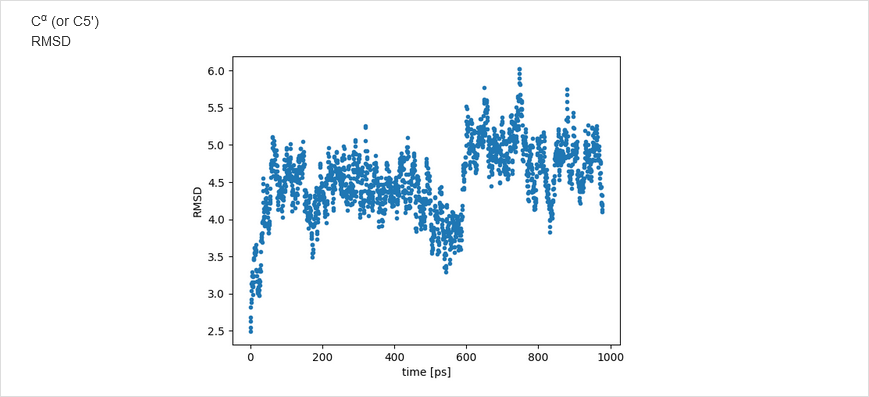
The NARES-minimized structure is visualized and superposed on the reference structure; the root mean square deviation
is displayed.
COARSE-GRAINED MOLECULAR DYNAMICS OF NUCLEIC ACIDS
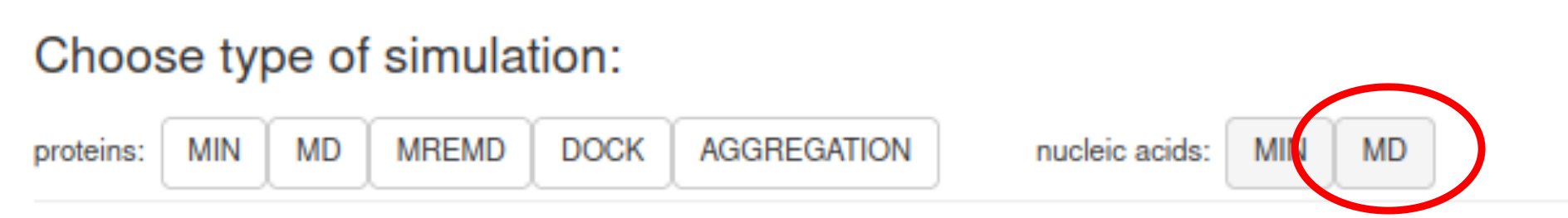
This tutorial covers running the coarse-grained molecular dynamics for nucleic acids with NARES-2P force field.To start, select MD in the nucleic acid section (on the right, as shown in the picture below).
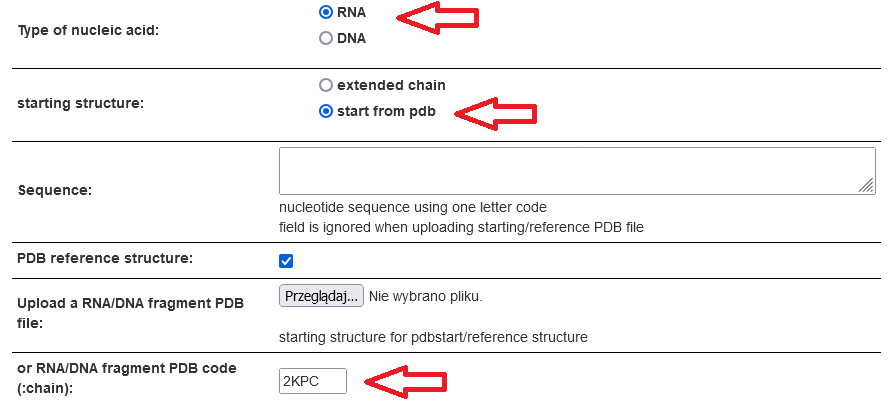
The simulation can be started from the extended/random structure or from the reference structure uploaded in the PDB
format or specified by the PDB code to download it from the PDB. The reference structure can also be input this way
even for extended/random starts. The sequence can either be input explicitly (in
one-letter code) or taken from the reference structure.
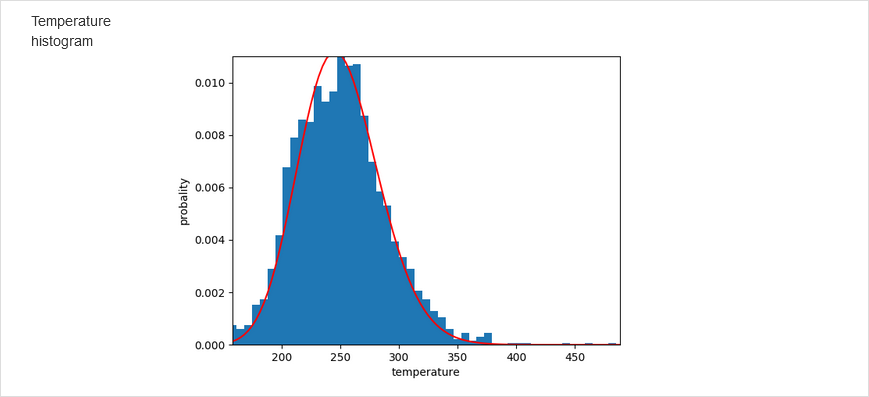
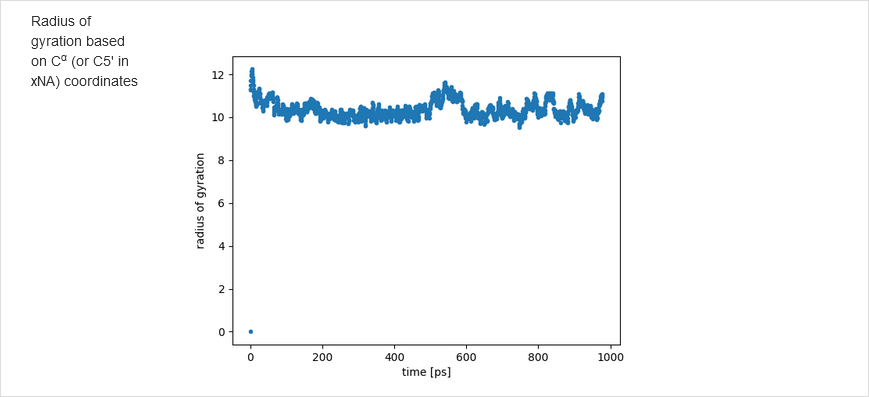
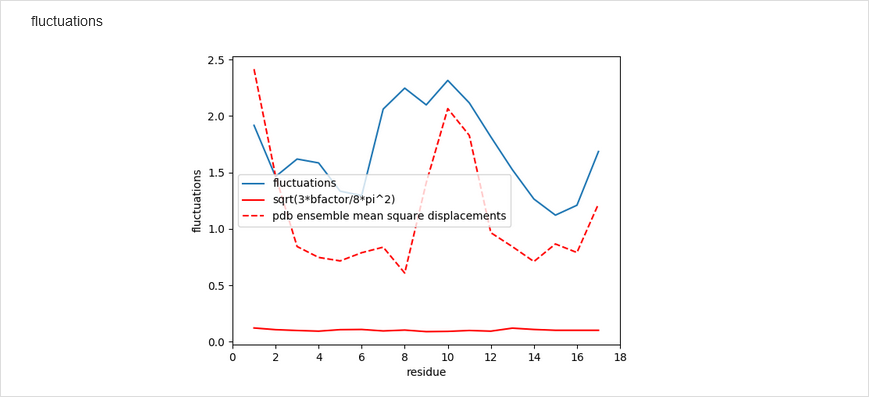
Subsequently, the number of time steps, time-step lenght, and bath temperature can be changed from the default values. Note that NARES-2P slightly underestimates melting temperature. Either the Berendsen or the Langevin thermostat can be selected. Having set the simulations, click Save & Submit. After the simulations are finished, the results are displayed in the graphical form. The graphs include temperature distribution, energy variation, residue fluctuations, which are compared with the B factors from crystal structure. The time-evolution of the molecule with time is also displayed as a movie.